|
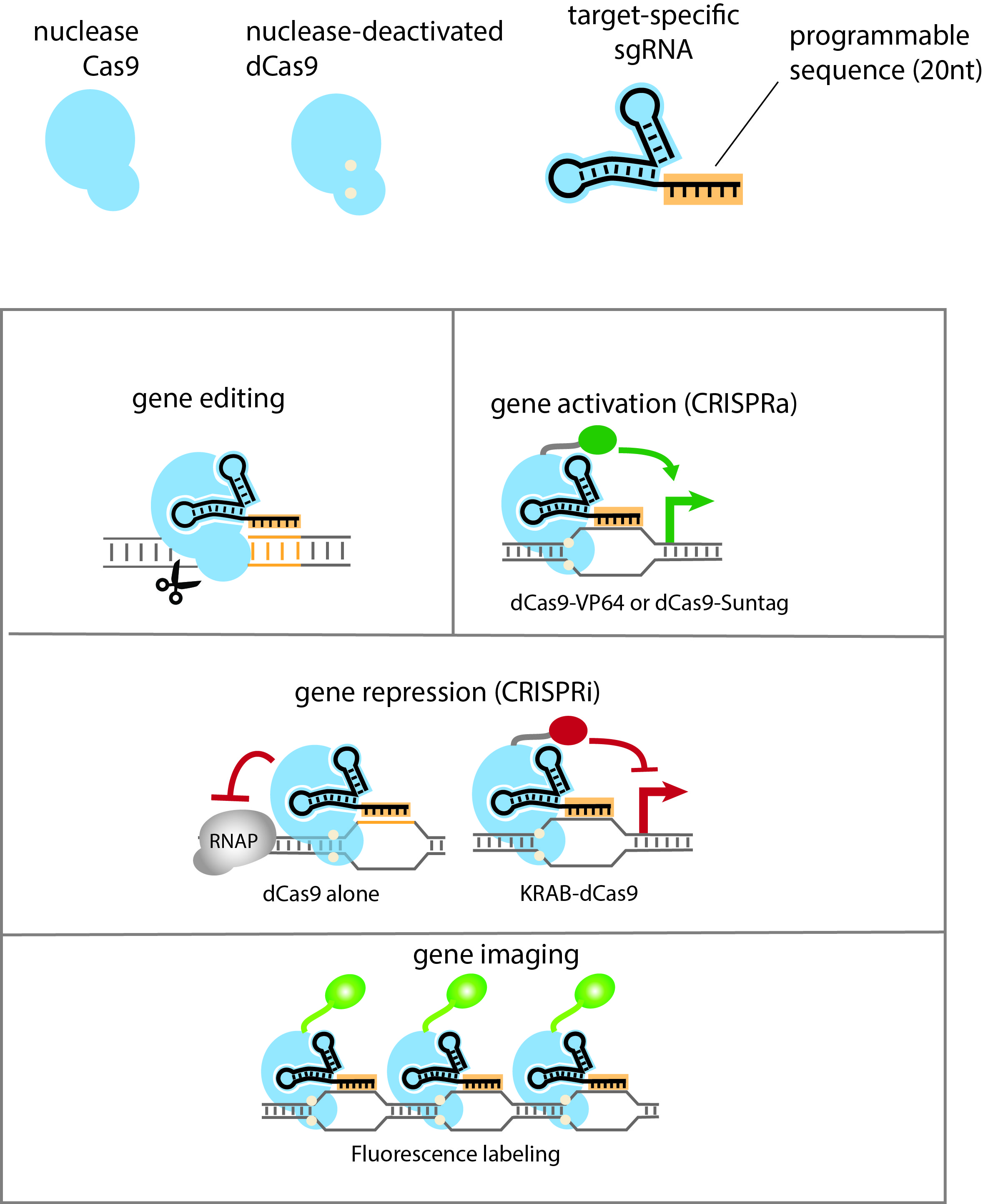
The bacterial antiviral system CRISPR (clustered regularly interspaced short palindromic repeats) has been developed as a flexible technology for multiple purposes of targeted genome engineering, including editing (modifying the genome sequence), gene repression (down-regulation) or activation (up-regulation of the target gene). The system is highly programmable, utilizing an invariant protein , - a nuclease Cas9 for editing or a nuclease-deficient dCas9 or dCas9 fusion protein for repression/activation, - and a custom-designed single guide RNA (sgRNA) for target DNA recognition (Ref). For the commonly used Streptococcus pyogenes CRISPR, target specificity is determined jointly by a 20-nt DNA-binding sequence on the sgRNA and a PAM sequence (protospacer adjacent motif, N(A/G)G) on the DNA.
|
|
| Reference: |
| 1 |
Chen, B. et al. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell 155, 1479-1491, doi:10.1016/j.cell.2013.12.001 (2013).
|
| 2 |
Gilbert, Luke A. et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159, 647-661, doi:10.1016/j.cell.2014.09.029 (2014).
|
| 3 |
Qi, Lei S. et al. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 152, 1173-1183, doi:10.1016/j.cell.2013.02.022 (2013). |
| 4 |
Tsai, S. Q. et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. (2014).
|
| 5 |
Wang, T., Wei, J. J., Sabatini, D. M. & Lander, E. S. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–4 (2014).
|
|

